File Upload Guide
Overview
FINDVIZ supports multiple fMRI file formats. This guide explains how to upload files and what formats are supported.
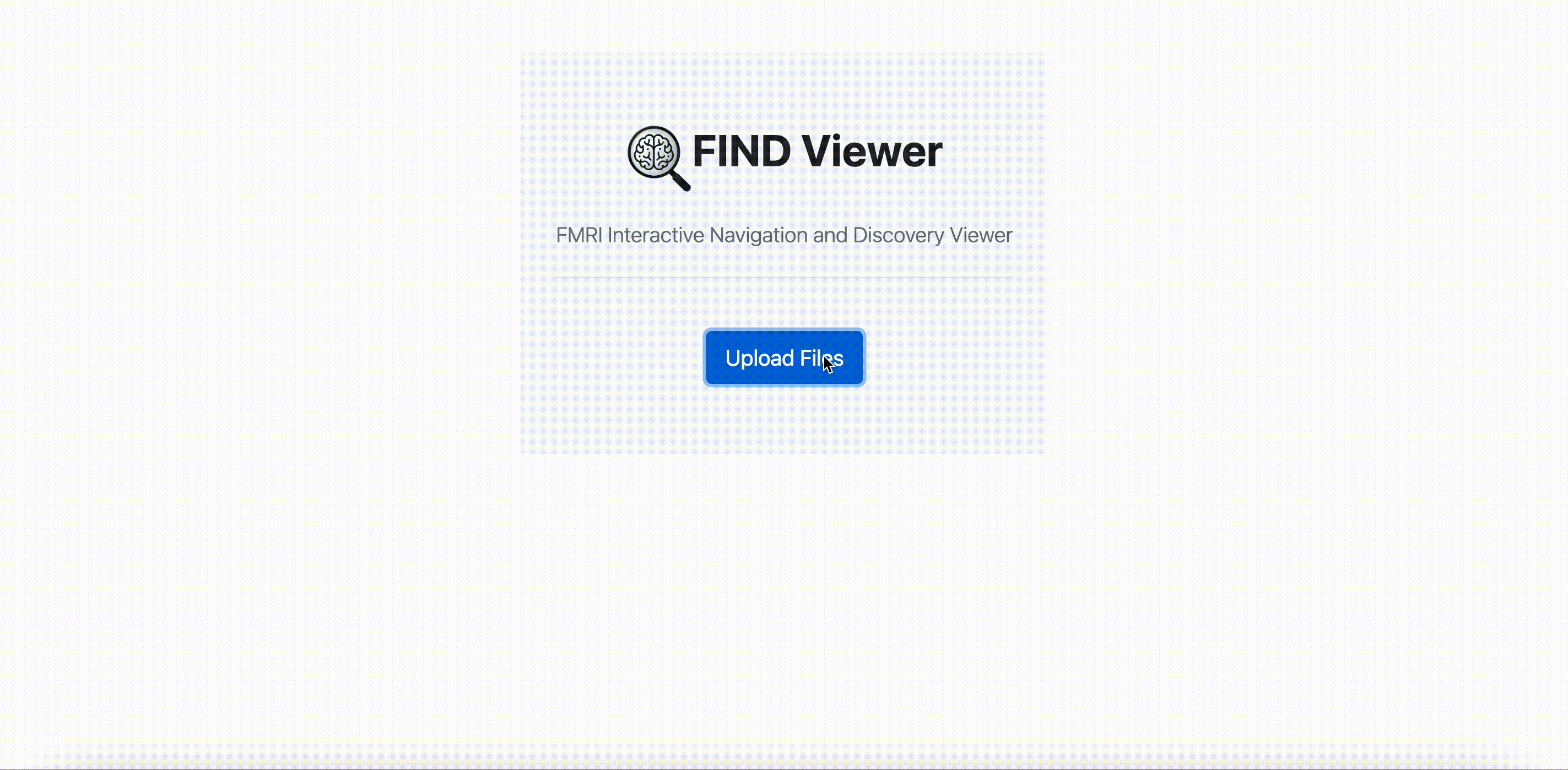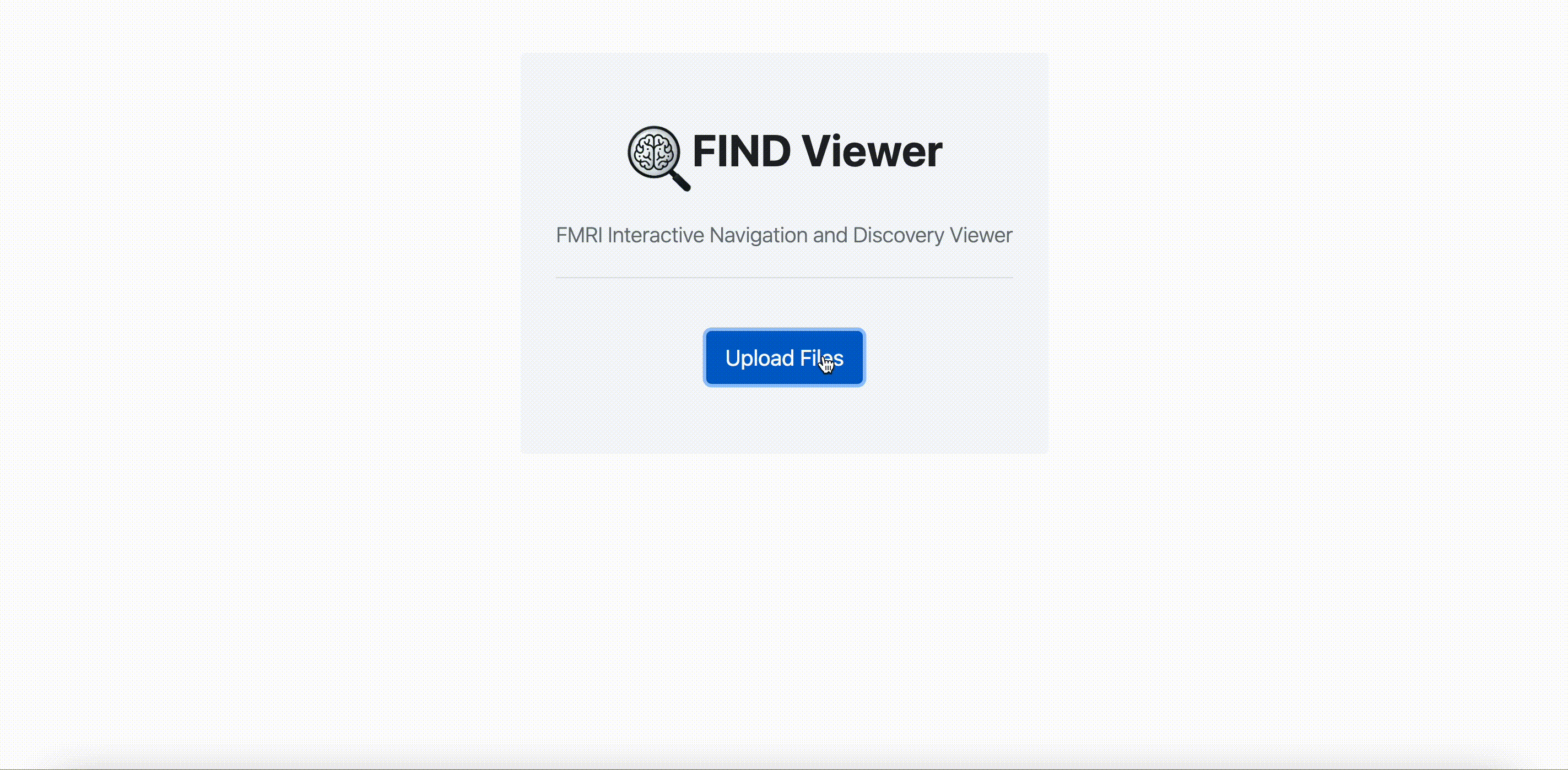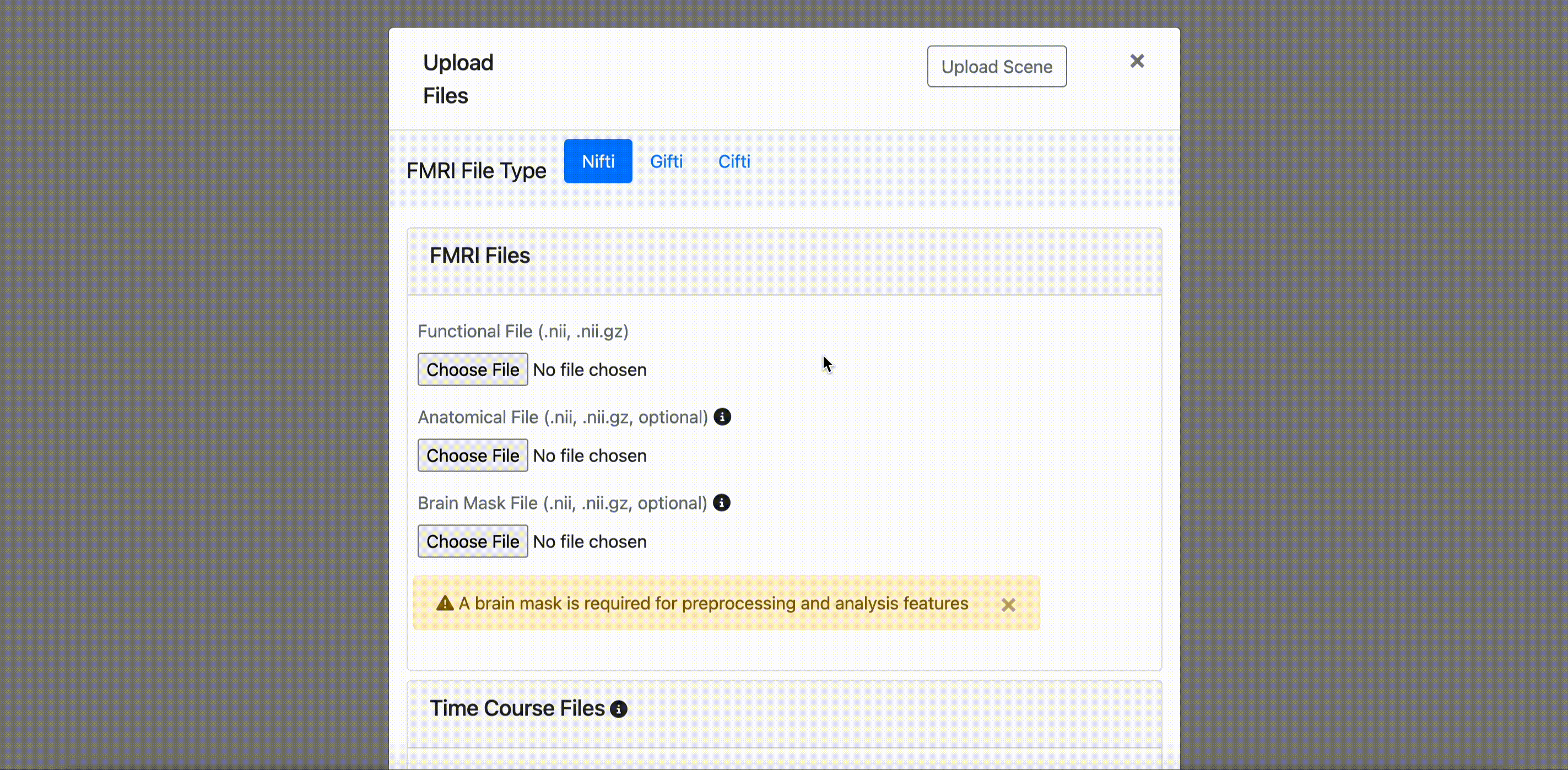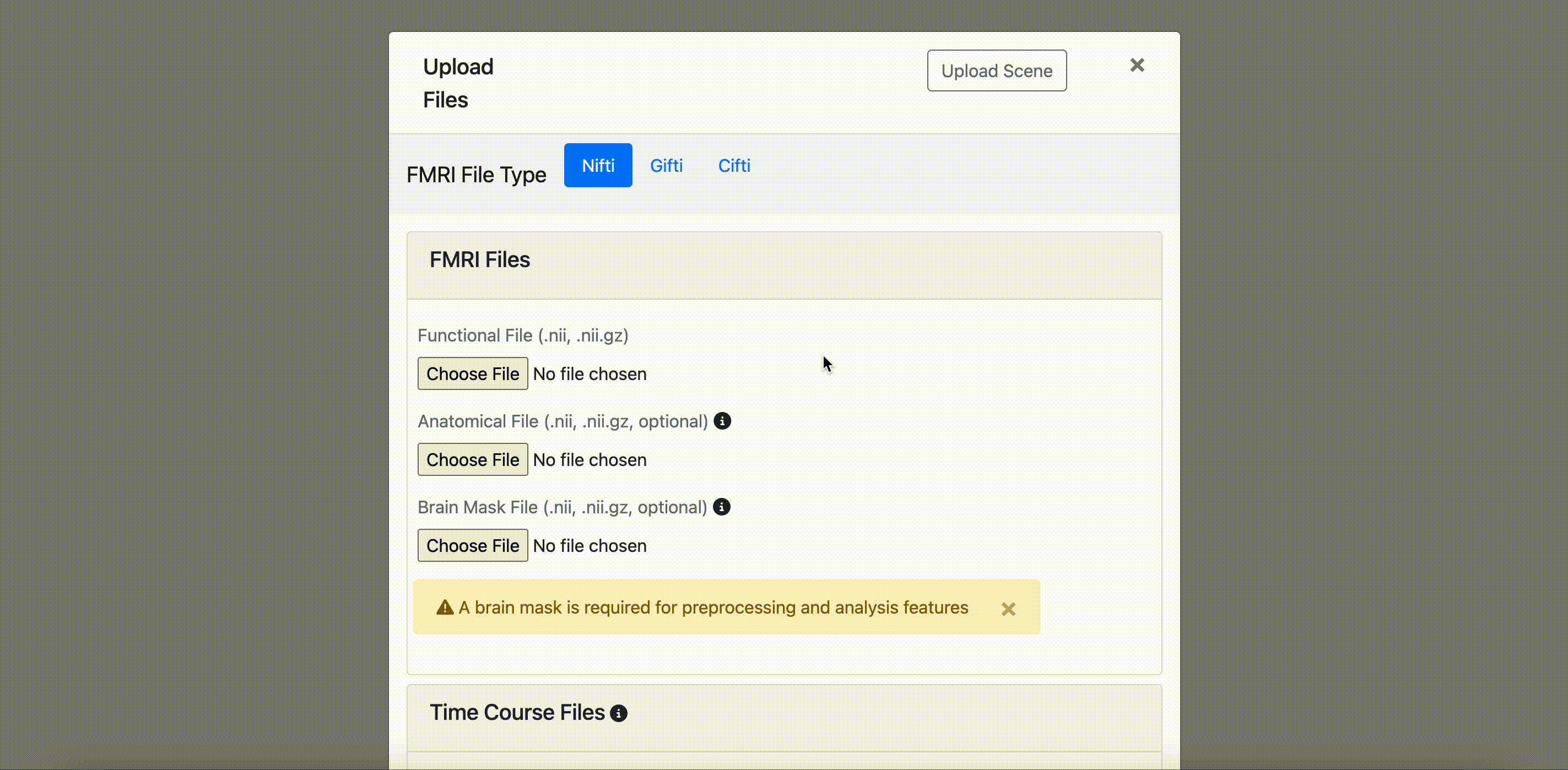
Table of Contents
FMRI File Formats
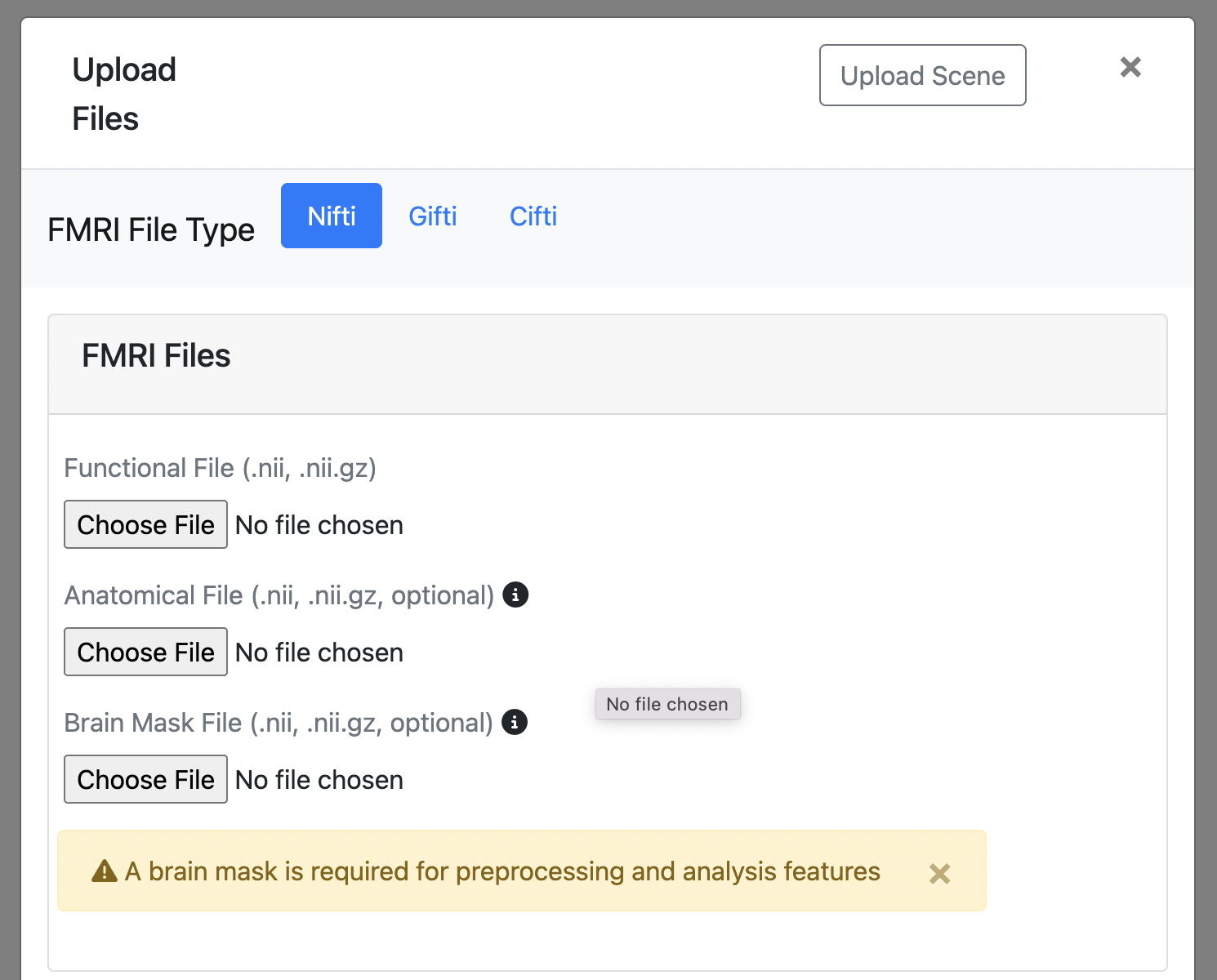
NIFTI (.nii, .nii.gz)
- Required Functional file: 4D functional brain images (.nii or .nii.gz)
- Optional Anatomical file: 3D structural brain images (.nii or .nii.gz)
- Optional Brain mask file: Binary masks for brain extraction (required for some analysis functions)
All preprocessing and analysis functions for nifti files require a brain mask.
Important Notes
- All NIFTI images must have the same voxel dimensions, even if in the same coordinate space (e.g. MNI152).
- A functional file is required, but you can visualize it without an anatomical file or mask.
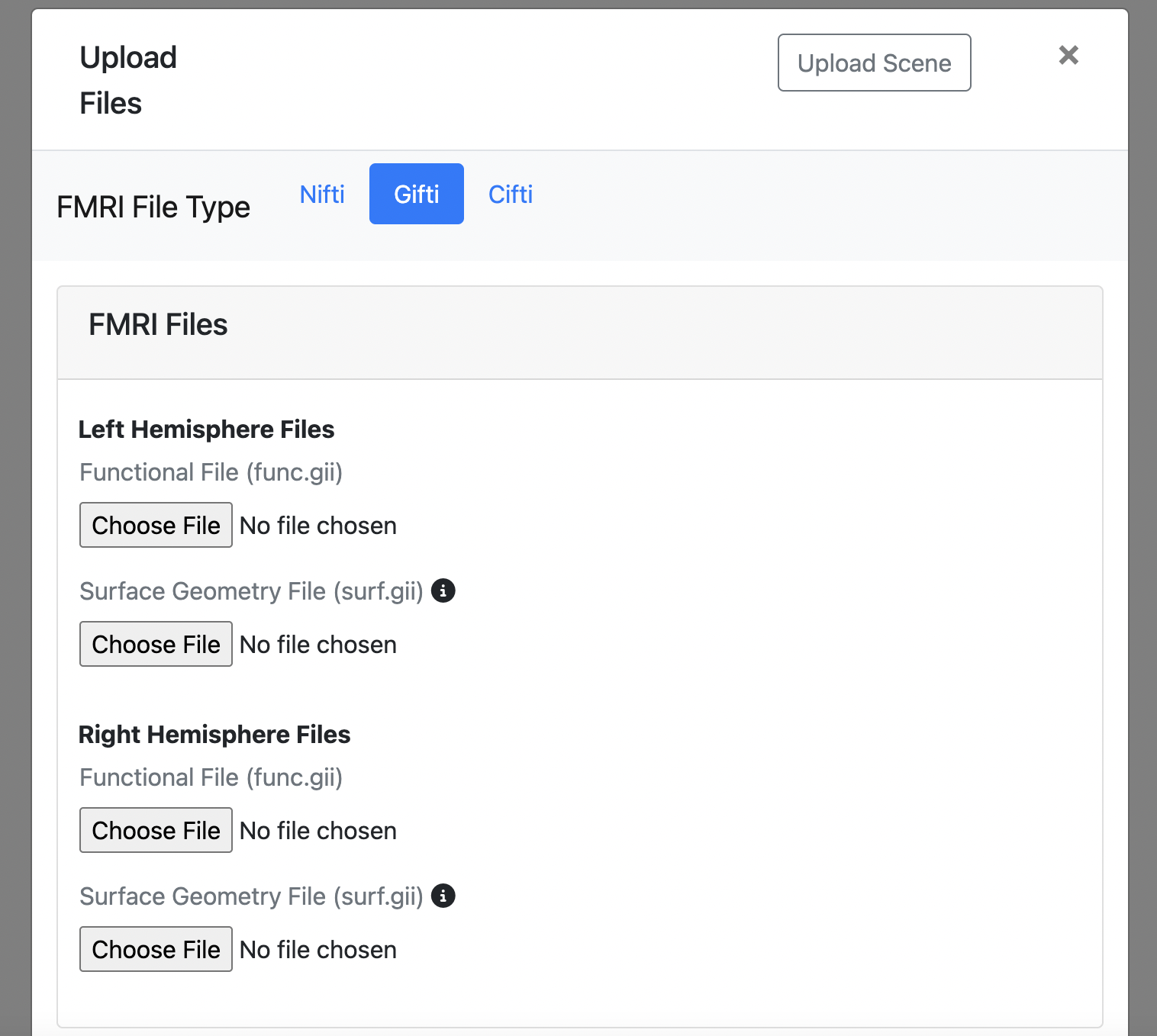
GIFTI (.gii)
- Required* Left hemisphere functional file: Surface-based functional data (func.gii)
- Required* Left hemisphere geometry file: 3D representation of the brain's outer surface (surf.gii)
- Required* Right hemisphere functional file: Surface-based functional data (func.gii)
- Required* Right hemisphere geometry file: 3D representation of the brain's outer surface (surf.gii)
Important Notes
- *You need either both left hemisphere files OR both right hemisphere files (functional and geometry). You can also upload all four files for complete visualization.
- Each hemisphere requires both its functional file and geometry file.
- Surface geometry files (surf.gii) provide the 3D mesh on which functional data will be displayed.
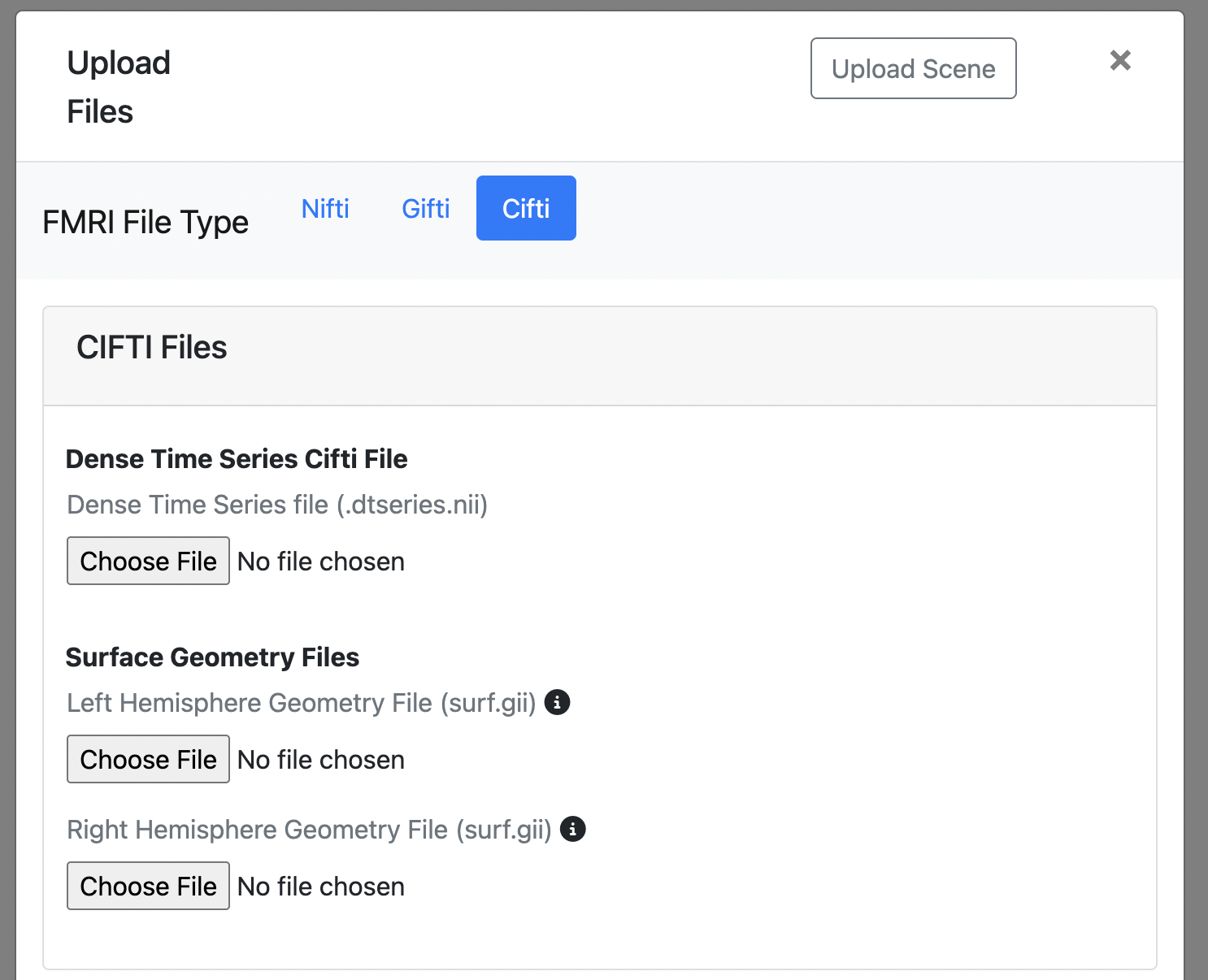
CIFTI (.dtseries.nii)
- Required Dense time series file: Combined surface and volume data (.dtseries.nii)
- Required* Left hemisphere geometry file: 3D representation of the brain's outer surface (surf.gii)
- Required* Right hemisphere geometry file: 3D representation of the brain's outer surface (surf.gii)
Important Notes
- *You need at least one hemisphere geometry file (either left or right). You can upload both for complete visualization.
- CIFTI files combine both surface and volume data into a single file. However, FINDVIZ currently only supports visualization of the surface data.
Time Course Files
In addition to fMRI data, FINDVIZ allows you to upload:
- Optional Time series files (.csv, .txt): Custom time course data for visualization alongside fMRI data
- Optional Task design files (.csv, .tsv): Experimental design matrices for visualization
Time Course Files
Upload .csv or .txt files containing time course data.
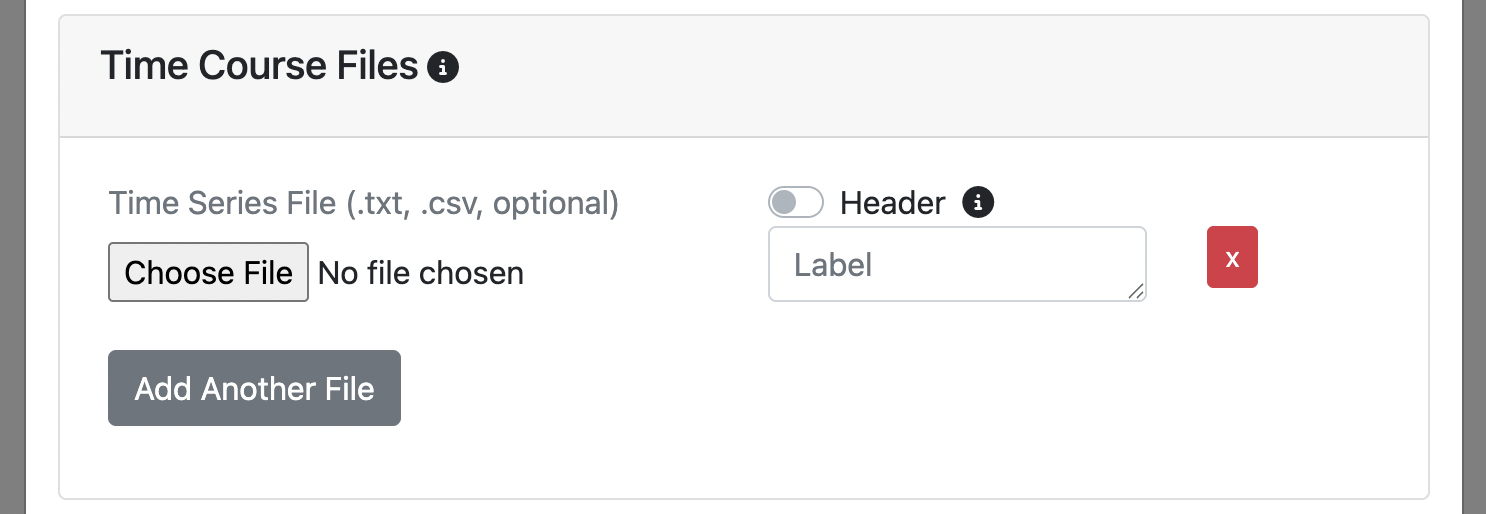

These files should:
- Have the same number of time points as your fMRI data. For concurrently recorded physiological signals, this typically means resampling/aligning your signals to the fMRI sampling times. Several open-source solutions in the Python ecosystem offer this functionality, e.g. NeuroKit2.
- Be arranged in one column
- Be temporally aligned with your fMRI data
Optional
- You can provide a custom label for the time course. If not specified, the display label will be the file name.
-
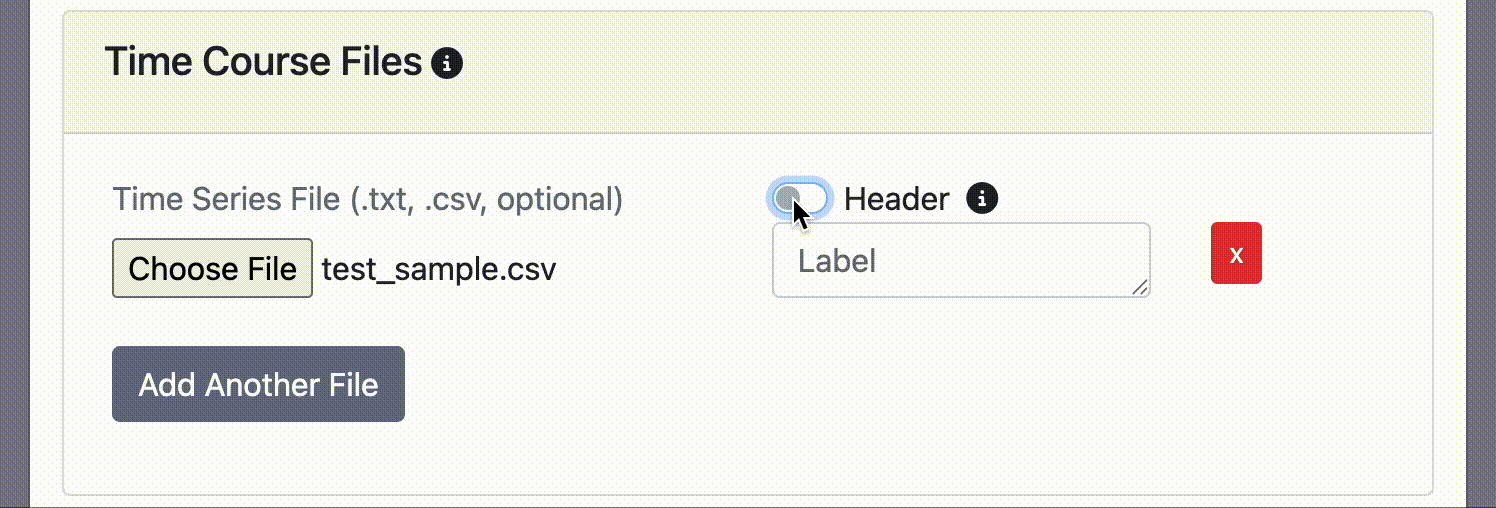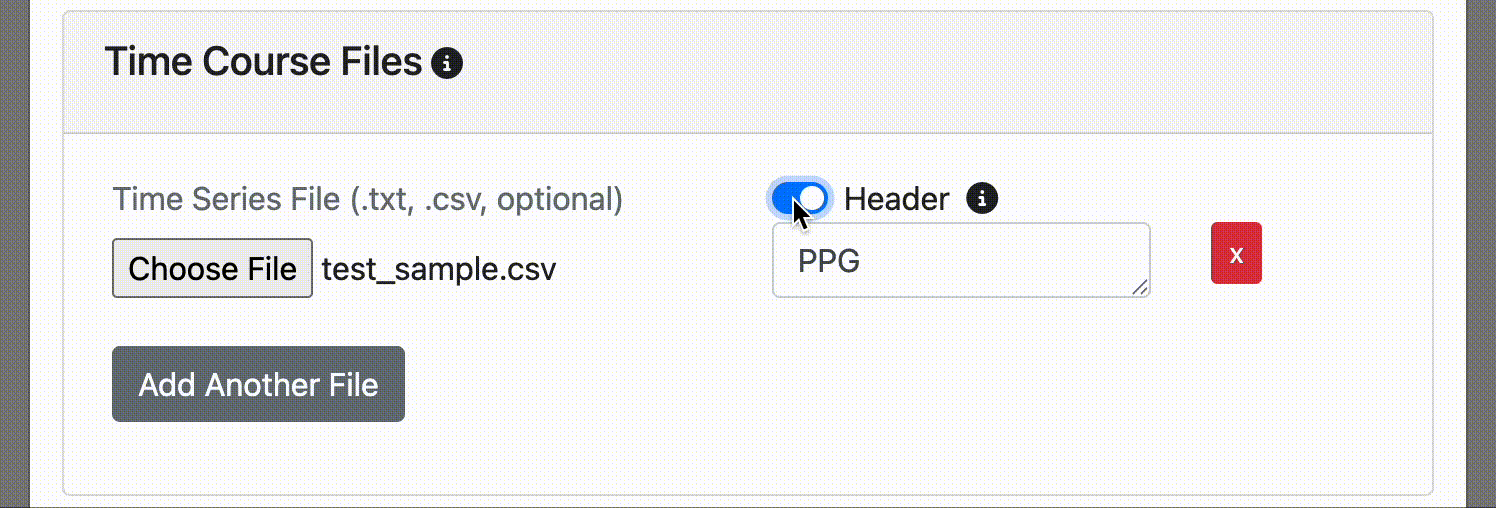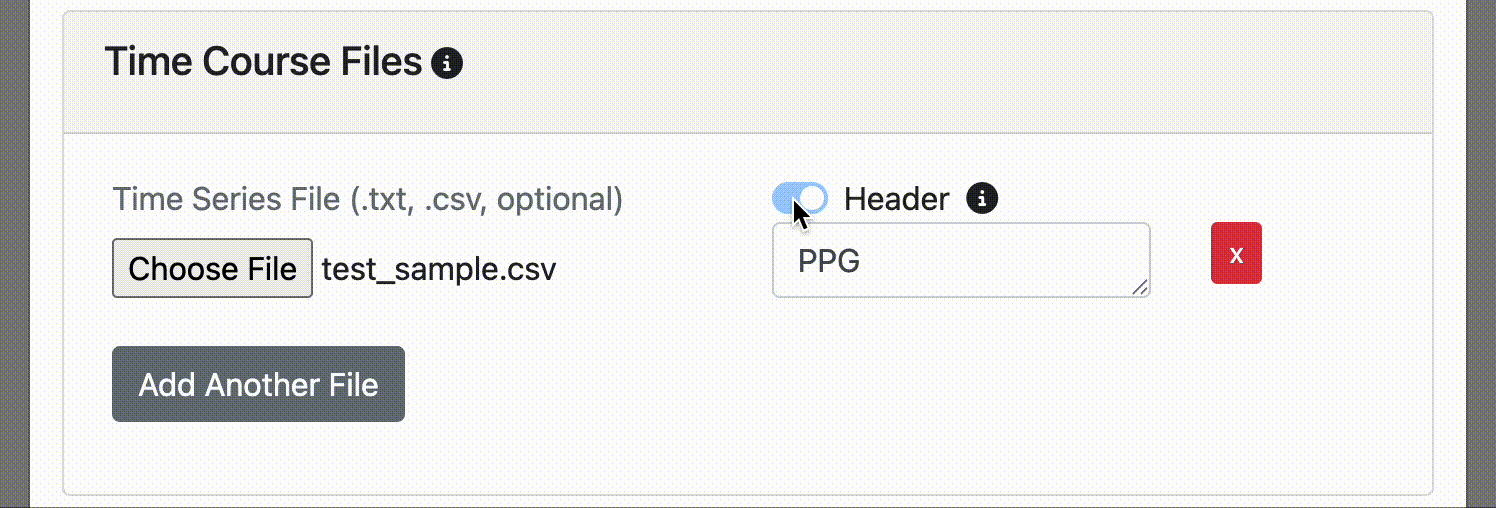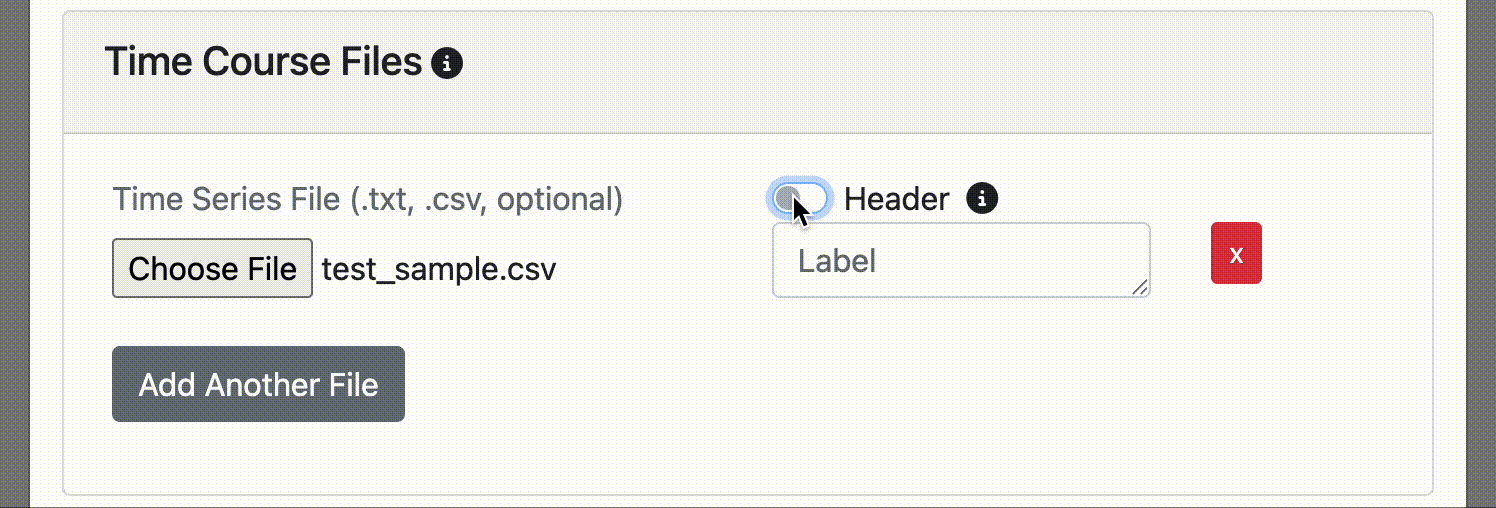
It's common to have a column label or header at the top of your time course file. If you do, simply click the header switch for FINDVIZ to recognize the custom label in the header. Note, uploading a time course file with a header will raise an error if the header option switch is not clicked on upload.
-
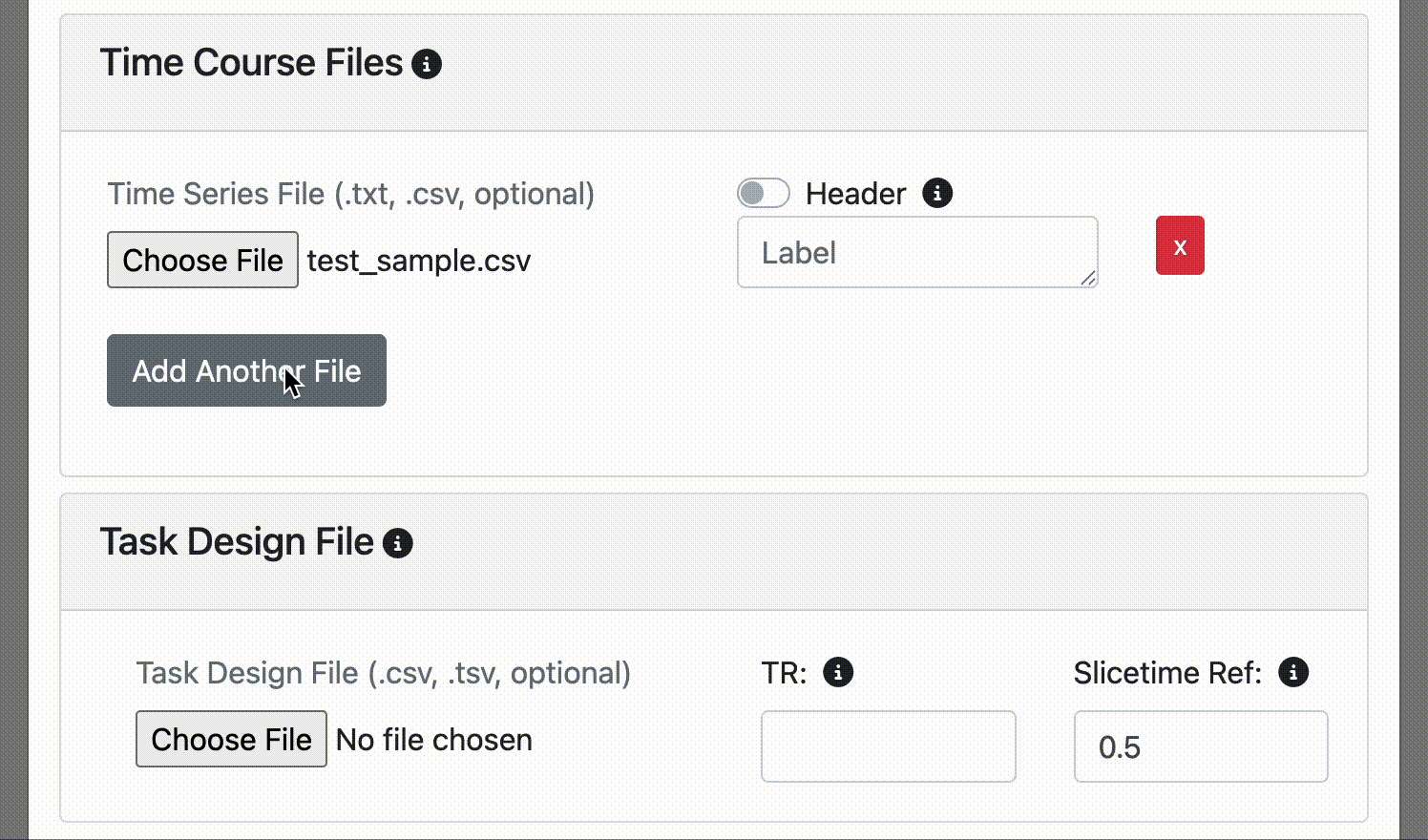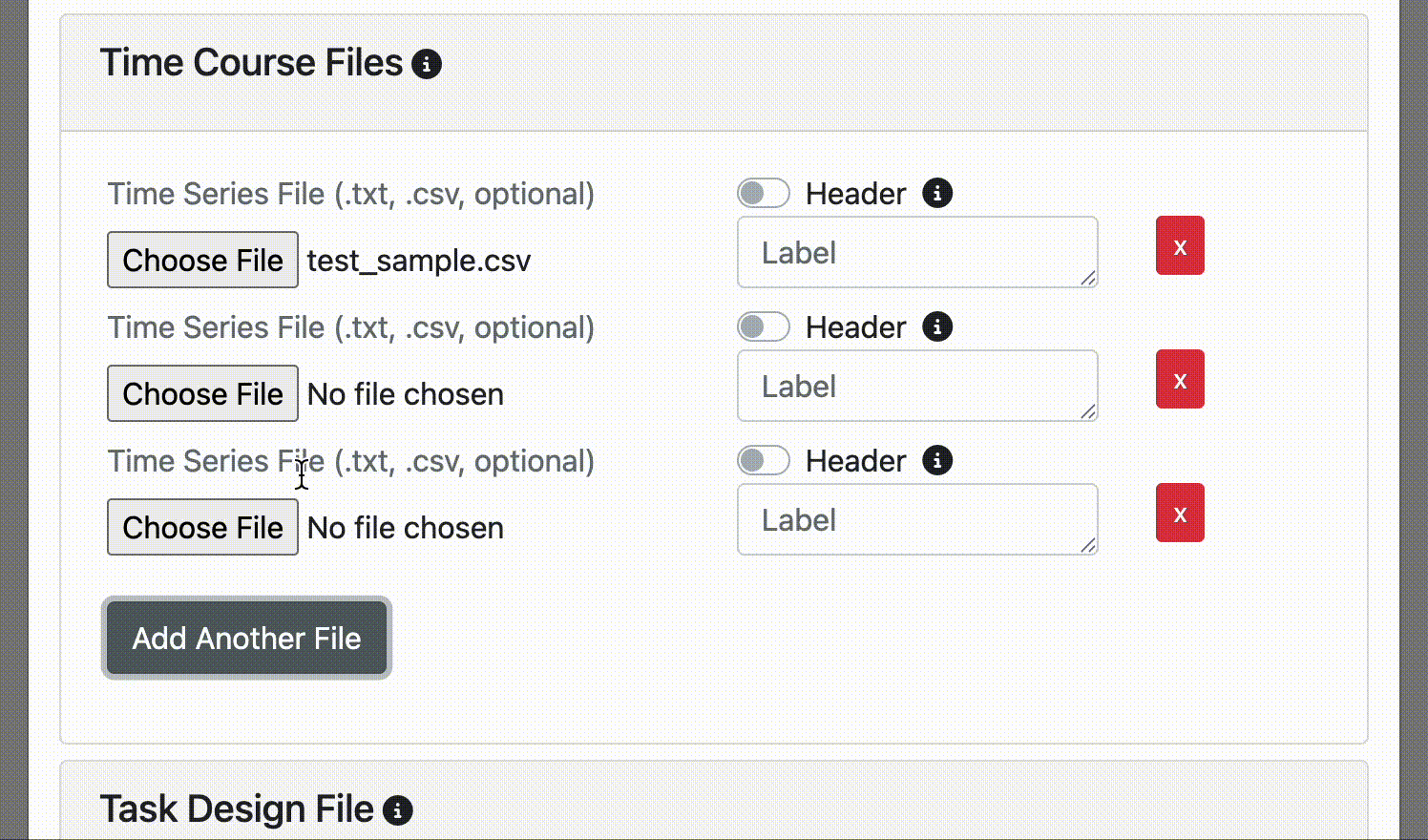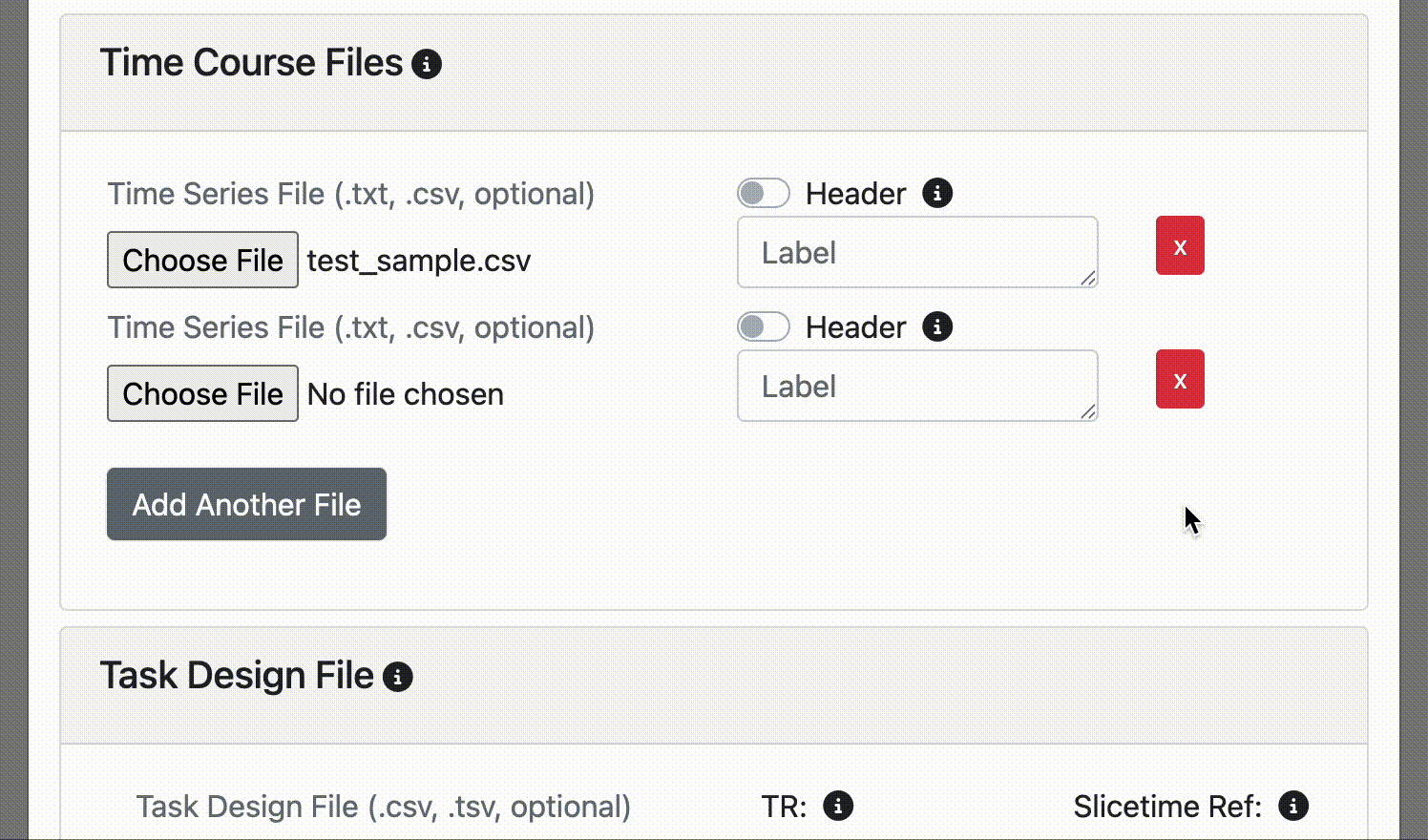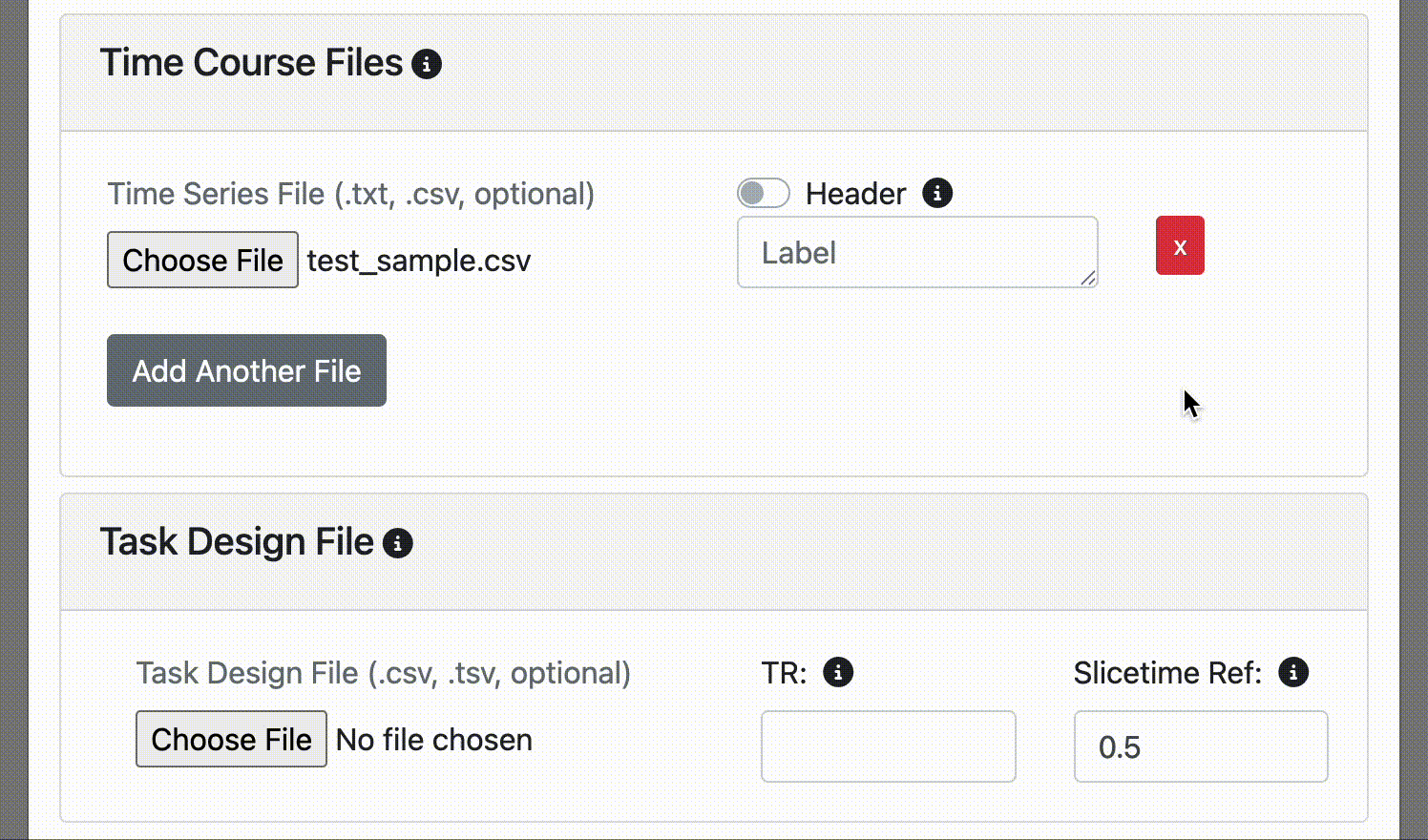
If you want to add another time course. Click the Add Another File button in the Time Course Files section.
Task Design Files
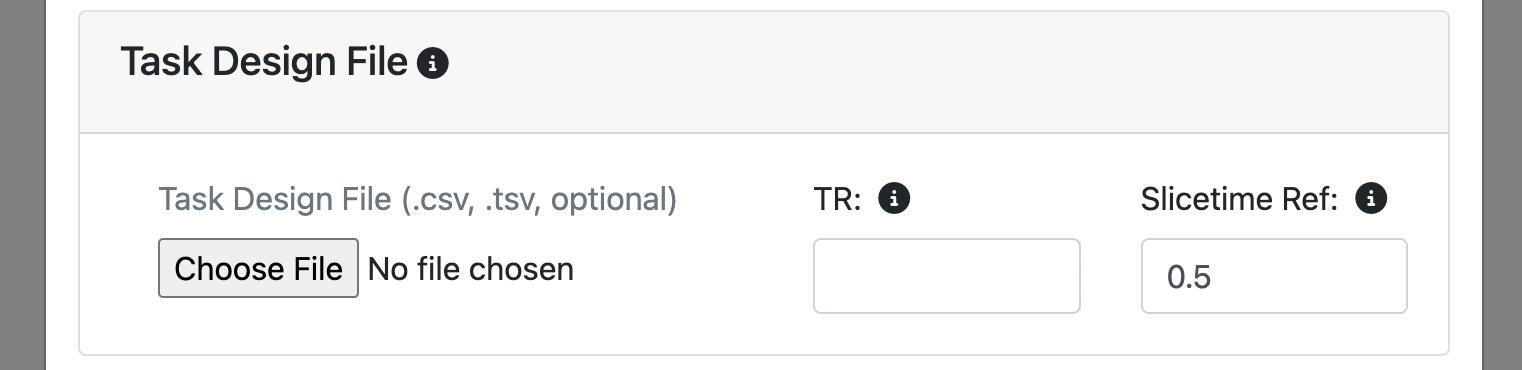
Upload a .csv or .tsv file in the Task Design File section.

The file must contain:
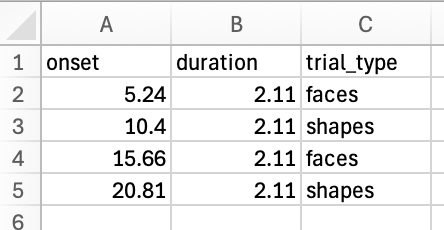
- An 'onset' column with values in seconds
- A 'duration' column with values in seconds
- Optionally, a 'trial_type' column for multiple event types
Example File:

Specify the TR (repetition time) of your fMRI data. This is needed for aligning the task events with the fmri data.
Specify the slicetime 'reference value' (default is 0.5) - the time of the reference slice used in the fmri slice timing correction algorithm. It is expressed as a percentage of the TR (repetition time): 0 - 1. For example, if slicetiming was referenced to the slice at the middle of the fmri acquisition, it would be 0.5. Default is 0.5, which is commonly used in fmri slice-timing algorithms.
Command Line Upload (Alternative)
Command Line Options
You can also upload files directly from the command line:
# Launch with NIFTI files
findviz --nifti-func func.nii.gz --nifti-anat anat.nii.gz
# Launch with GIFTI files (all four files)
findviz --gifti-left-func left.func.gii --gifti-right-func right.func.gii --gifti-left-mesh left.surf.gii --gifti-right-mesh right.surf.gii
# Launch with GIFTI files (left hemisphere only)
findviz --gifti-left-func left.func.gii --gifti-left-mesh left.surf.gii
# Launch with GIFTI files (right hemisphere only)
findviz --gifti-right-func right.func.gii --gifti-right-mesh right.surf.gii
# Launch with CIFTI files (both hemispheres)
findviz --cifti-dtseries data.dtseries.nii --cifti-left-mesh left.surf.gii --cifti-right-mesh right.surf.gii
# Launch with CIFTI files (left hemisphere only)
findviz --cifti-dtseries data.dtseries.nii --cifti-left-mesh left.surf.gii
# Add time series data
findviz --nifti-func func.nii.gz --timeseries timeseries1.csv timeseries2.csv
# Add task-design data
findviz --nifti-func func.nii.gz --task-design task.csv --tr 2.0
--slicetime-ref 0.5
Developer Notes
FINDVIZ uses the Python neuroimaging library nibabel to read and write neuroimaging files.