Preprocessing Guide
Overview
FINDVIZ offers several preprocessing options for your fMRI and time course data. These tools are not meant to serve as a complete preprocessing pipeline, but are designed to facilitate enhanced visualization and pattern discovery. See our data preparation section for more information on preprocessing your data before uploading to FINDVIZ.
Table of Contents
Available Preprocessing Methods
Preprocessing Methods
Preprocessing tools can be accessed from the fMRI and time course preprocessing panels in the interface.
FMRI Panel
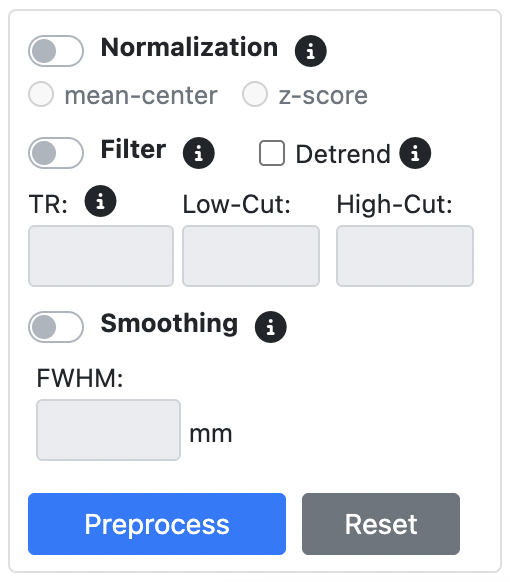
Timecourse Panel
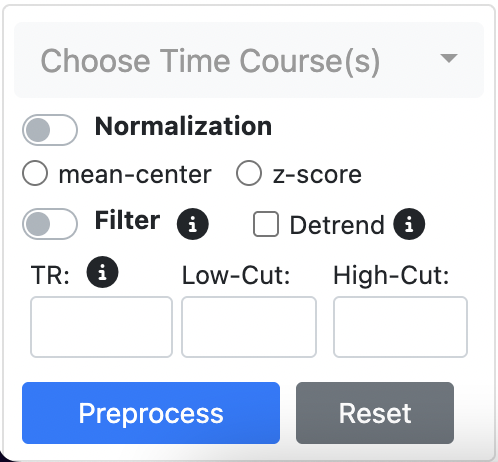
-
Linear Detrending
- Removes linear trends from fMRI or time series data using a linear least-squares fit.
- Use when: Your data shows gradual drift over time that could obscure patterns of interest
- Implementation: linear detrending of time courses is performed using SciPy (v1.13.1) detrend algorithm
-
Time Series Normalization
- Mean-center or z-score fMRI or time series data using a linear least-squares fit.
- Use when: You need to standardize signal amplitudes across different regions, or when comparing time series with different units or scales
- Implementation: mean-centering and z-scoring is perfomed with custom Python algorithms.
-
Temporal Filtering
- Band-pass, low-pass or high-pass fmri or time series data.
- Parameters: TR (repetition time of fMRI data), low-cut in hertz (hz), high-cut in hertz(hz). Specify only high-cut to low-pass filter (filter out frequencies above the cut-off). Specify only low-cut to high-pass filter (filter out frequencies below the cut-off). Specify high-cut and low-cut to bandpass filter (filter out frequencies outside the frequencies of the low-cut, high-cut range).
- Use when: You want to isolate specific frequency components of your signal, such as removing high-frequency noise or low-frequency drift, or focusing on a specific frequency band of interest (e.g., 0.01-0.1 Hz for resting-state fMRI)
- Implementation: Temporal filtering is performed using Nilearn's (v0.10.4) Butterworth filter function (5th order), which applies a zero-phase forward and backward filter to preserve timing relationships in the data
-
Spatial Smoothing
Surface smoothing of CIFTI/GIFTI surface plots are not currently supported.
- Gaussian spatial filtering of NIFTI fMRI images
- Parameters: FWHM - full-width at half maximum, in millimeters
- Use when: You want to increase signal-to-noise ratio of your NIFTI images.
- Implementation: Spatial smoothing is performed using Nilearn's (v0.10.4) smooth_img function.
Order of Processing
When multiple preprocessing methods are selected, they are applied in the following order:
- Linear Detrending
- Temporal Filtering
- Time Series Normalization
- Spatial Smoothing
Important Considerations
Key Points to Remember
- Brain Mask: For NIFTI data, a brain mask is required for all preprocessing operations. If you didn't upload a mask, all preprocessing options are disabled.
- Reversibility: after completion of preprocessing, preprocessed data is directly displayed in the plot (time course or fMRI plots). Only one preprocessed state is stored in FINDVIZ at a time. To restore the original data you can click the Reset button.
Limitations
Remember that FINDVIZ is primarily a visualization tool. For comprehensive preprocessing of fMRI data, consider using a dedicated fMRI preprocessing software